RESEARCH
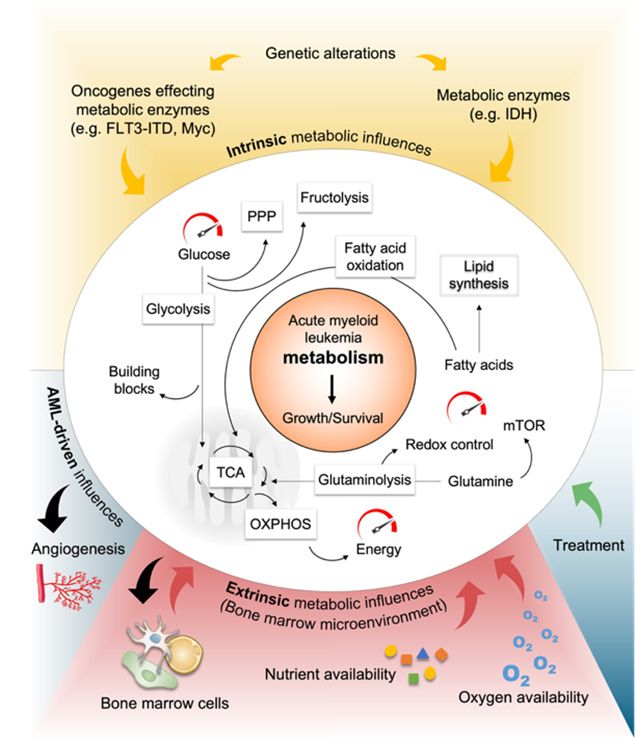
Metabolism of Acute Myeloid Leukemia
In this context, we are particularly interested in the question how changes in metabolic and signaling circuits make leukemic cells more or less responsive to environmental stimuli (e.g. nutrients, cell-cell interactions, cytotoxic drugs, hypoxia).
To do this, we are using cutting-edge technologies such as CRISPR/Cas9 screening, prime editing and endogenous tagging using CRISPR/Cas9, flow cytometry and fluorescence-activated cell sorting (FACS) and in cooperation mass spectrometry- and nuclear magnetic resonance (NMR)-based metabolomics. These techniques enable us to quickly identify genetic determinants of gene expression, proliferation and survival, leukemogenicity and metabolic changes in AML cell lines and AML patient derived blasts with the goal of discovering novel clues for AML therapy.
Patient-specific/individualized/pre-therapeutic
prediction of therapy sensitivities
In the Serve group, we have established a standardized, FACS-based application of BH3 profiling to measure heterogeneous patient material, leading to more patient-focused medicine.
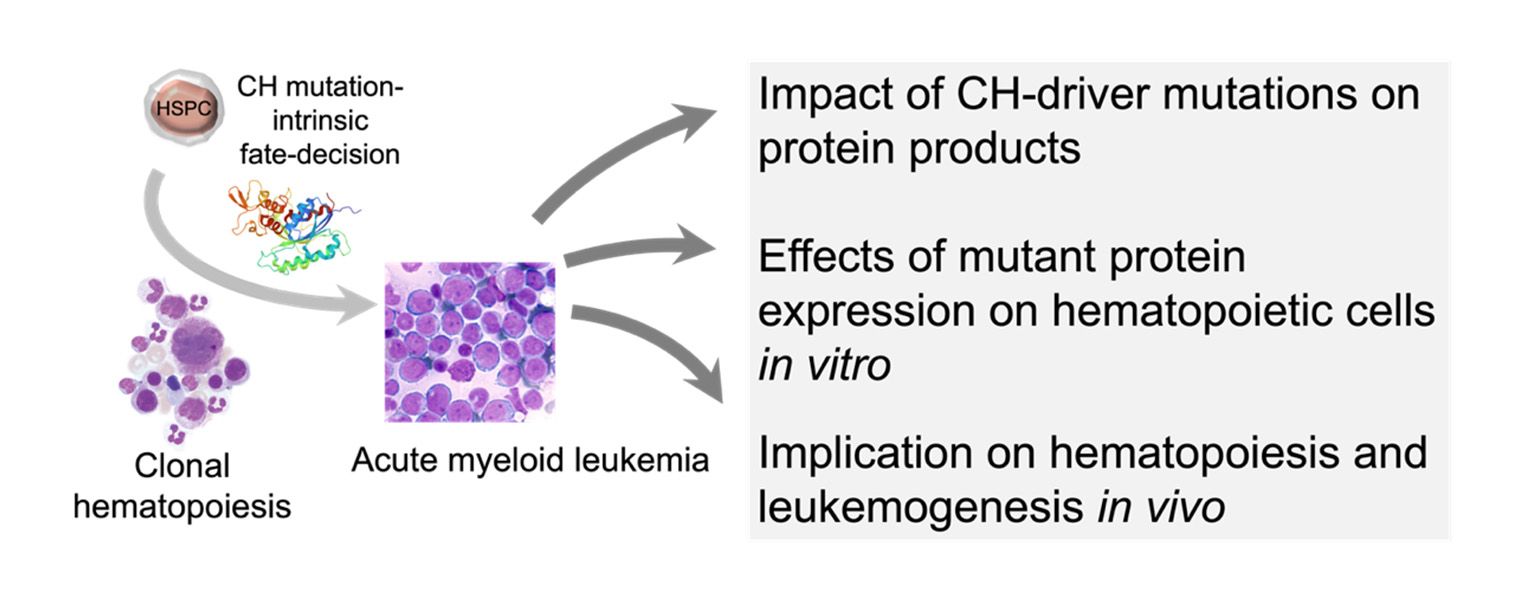
Comparative functional characterization of typical CH mutations in cardiovascular diseases and myeloid neoplasms
Molecular functions of amino acid sensing and processing
In the tumor microenvironment, cancer cells undergo metabolic adaptations that allow them to rapidly respond to dynamic environmental cues to meet their demands. This ability to adapt, which we currently refer to as metabolic flexibility, has been shown particularly in cells originating from Acute Myeloid Leukemia (AML).
Therefore, our research explores the molecular mechanisms by which cells can sense and respond to fluctuations in available metabolites.
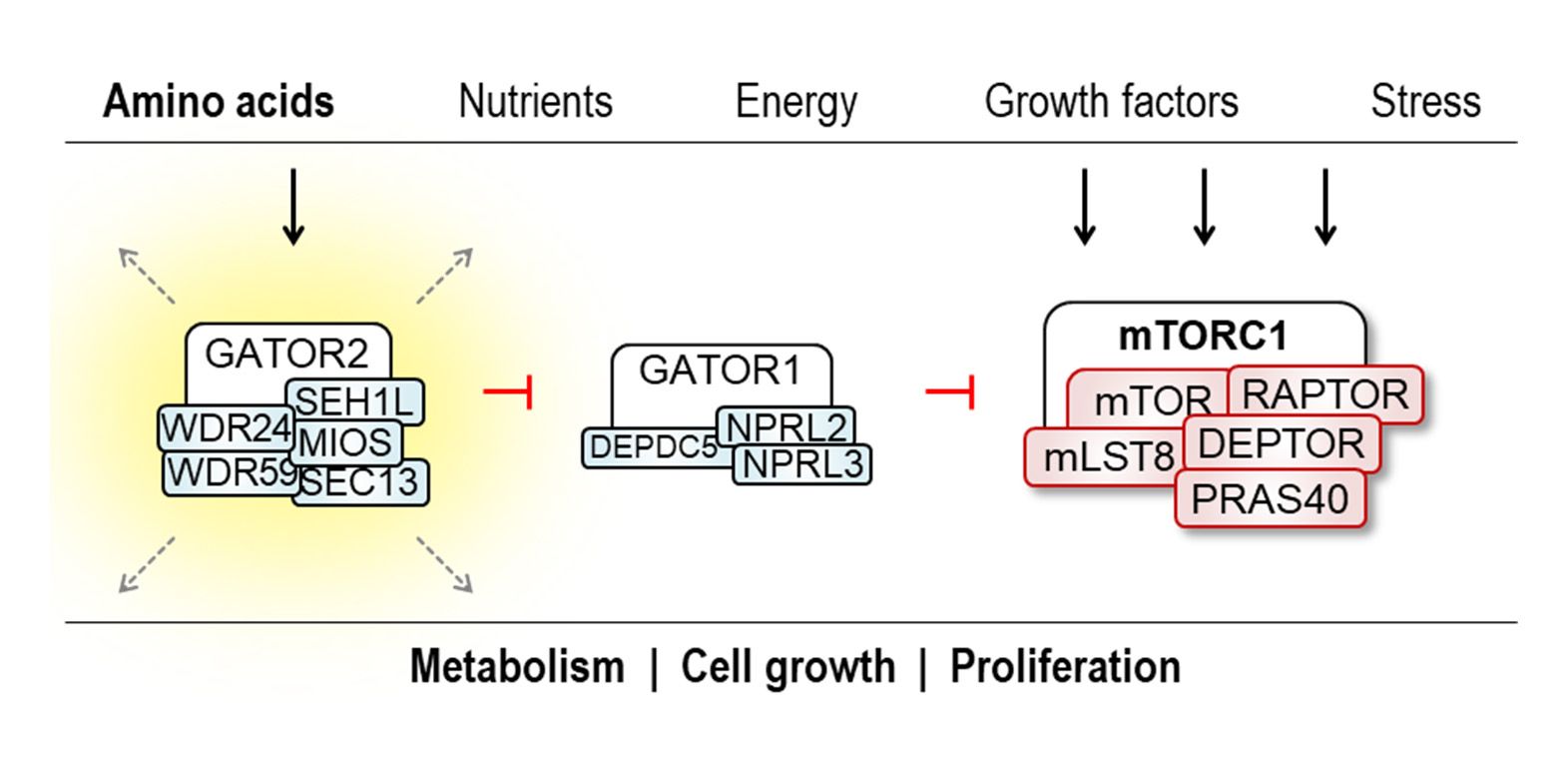
In the last decade, the discovery of two GATOR complexes has significantly advanced our understanding of cellular amino acid sensing and processing mechanisms. Amino acids play a crucial role in cell proliferation as building blocks for proteins, and thus sensing of amino acids is critical for cancer cells. This underscores the indispensable role of specialized sensor protein functions, including those of GATOR1 and GATOR2, to monitor the cellular amino acid pool.
GATOR2 communicates with different cellular signaling pathways, notably with the central metabolic hub, mTORC1, thereby exerting precise control over cellular growth. In this project, we employ endogenous protein tagging using CRISPR/Cas9 technology, along with co-immunoprecipitation (CoIP) and mass spectrometry (MS) approaches, to uncover previously unexplored protein interactions and functions. Our primary goal is to extend the functional repertoire of GATOR2 and to deepen the understanding of amino acid sensing by the GATOR2 complex using insights gained from its molecular architecture. Consequently, our findings not only deepen the understanding of the GATOR2 network but also offer promising avenues for the development of targeted therapeutic strategies.