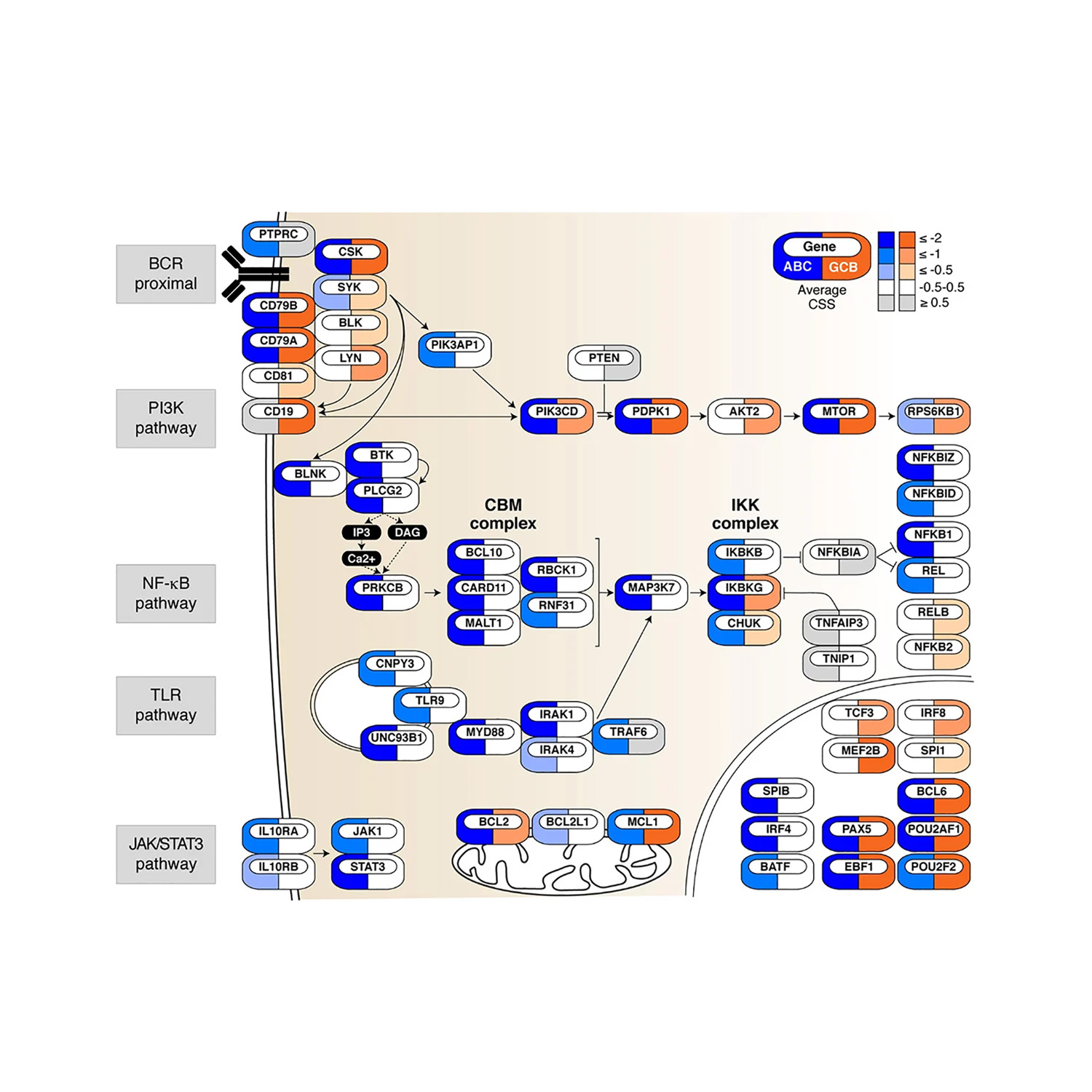
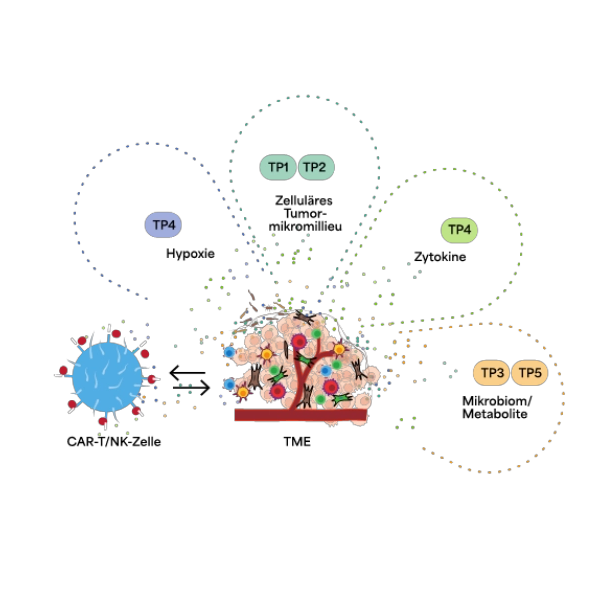
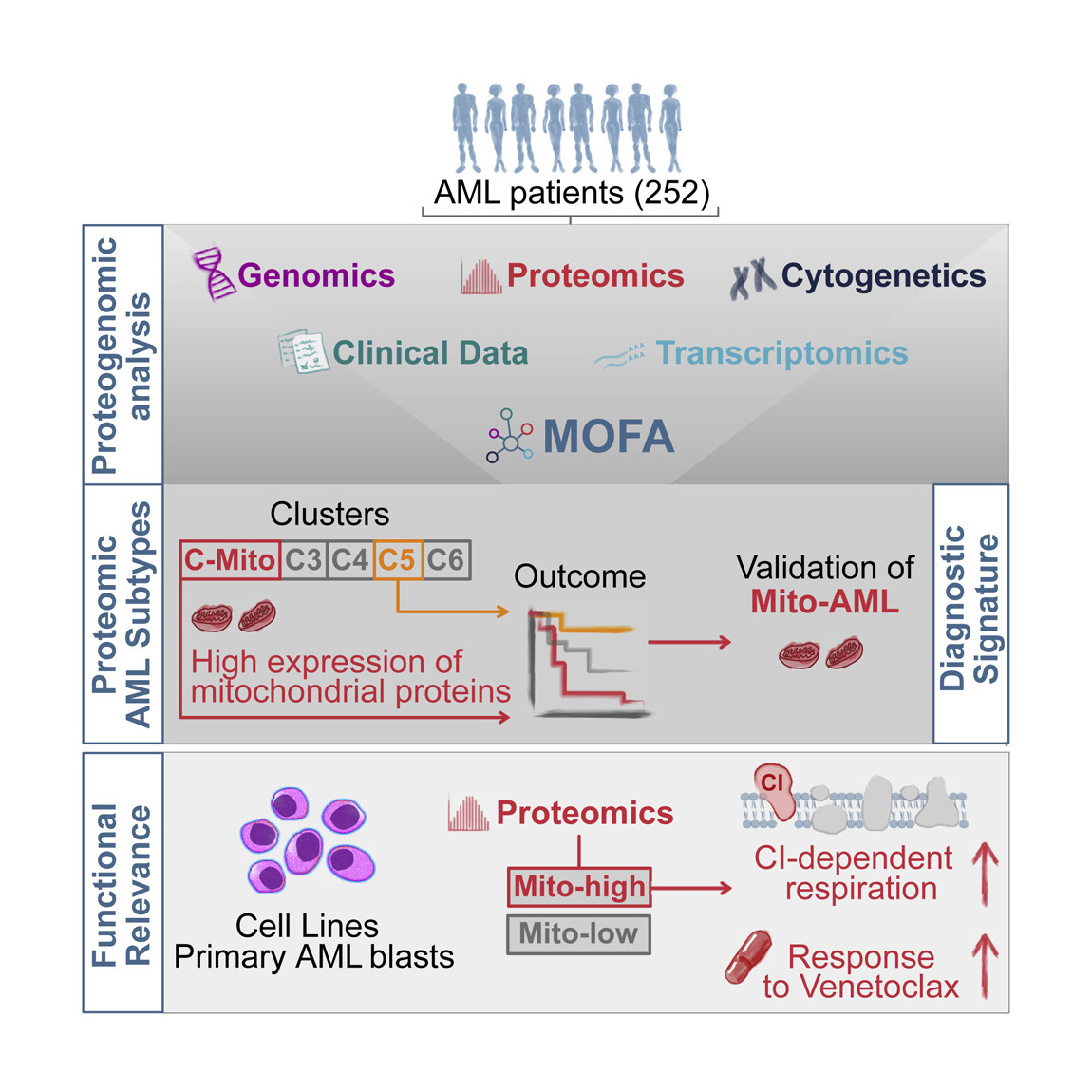
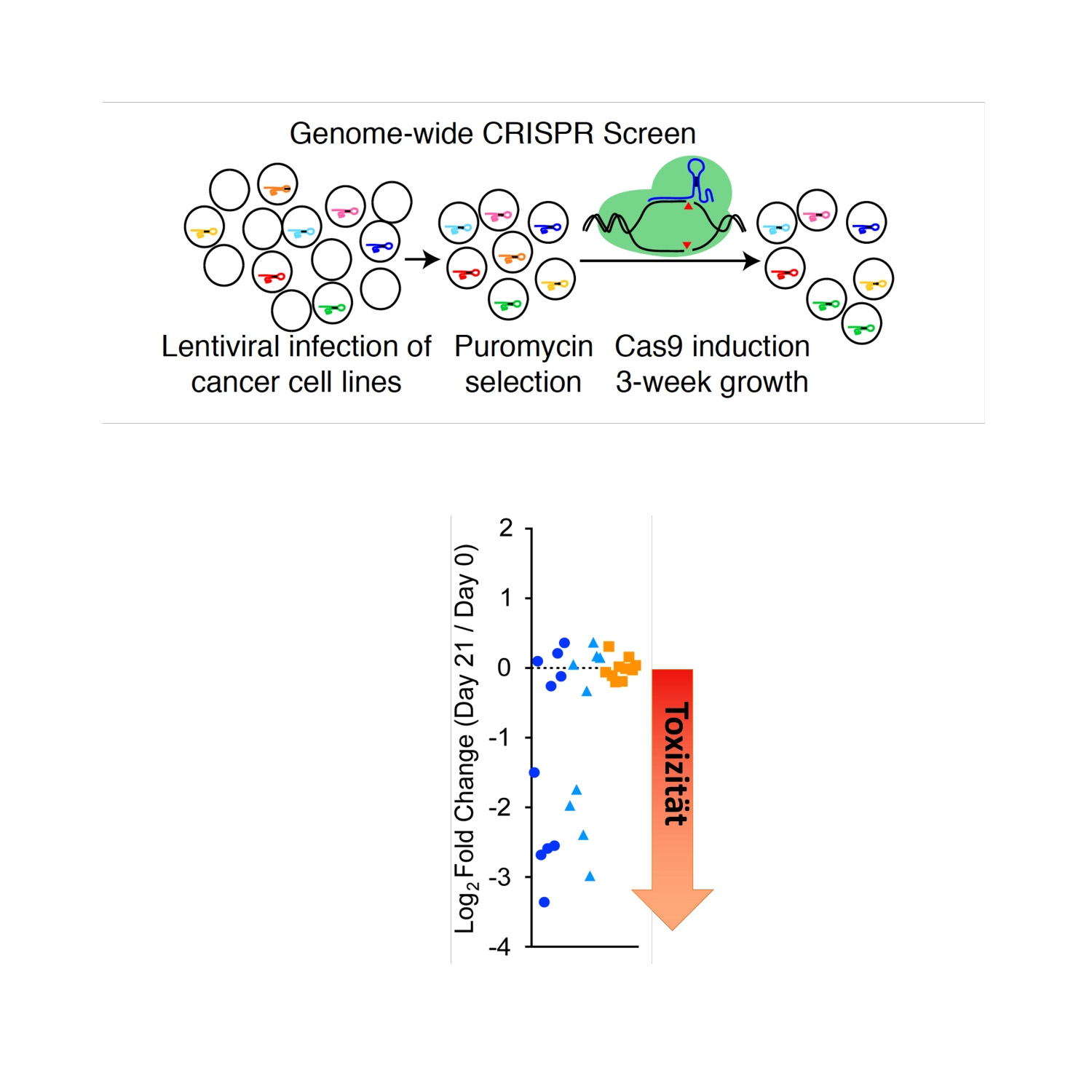
Privacy policy
We respect your personal data!
In the following, we describe how personal data is processed within the scope of our websites, functions and content as well as external offers (such as social media). We also describe what types of personal data are processed, to what extent and for what purpose.
1. Name and address of the data controller
The data controller within the meaning of the General Data Protection Regulation and other national data protection laws of the member states as well as other data protection regulations is:
Johann Wolfgang Goethe-Universität Frankfurt am Main
Theodor-W.-Adorno-Platz 1
60323 Frankfurt am Main
Mailing-address:
Goethe-Universität Frankfurt am Main
60629 Frankfurt
Telephone: +49-69-798-0 | Fax: +49-69-798-18383
Web: www.uni-frankfurt.de
The responsible person for the research content is Prof. Dr. Thomas Oellerich.
2. General information on data processing
2.1. Scope of the processing of personal data
When you contact us as an interested party, we collect personal data. This happens when you contact us by e-mail or telephone or when you use the contact form in the context of existing business relationships.
In these contexts, we process personal data, such as first and last name, address, email address, and telephone number.
2.2. Legal basis for the processing of personal data
If a processing of personal data is based on the consent of the data subject, Art. 6 (1) (a) of the EU General Data Protection Regulation (GDPR) serves as the legal basis. If personal data are processed in order to fulfill a contract with the data subject, Art. 6 (1) (b) GDPR serves as the legal basis. This also applies to processing operations that are necessary to carry out pre-contractual measures. If we process personal data in order to fulfill a legal obligation, Art. 6 (1) c GDPR serves as the legal basis. If the vital interests of the data subject or another natural person make it necessary to process personal data, Art. 6 (1) (d) GDPR serves as the legal basis. If we process data to protect the legitimate interests of our company or a third party and the interests, fundamental rights and freedoms of the data subject are not more important, Art. 6 (1) f GDPR serves as the legal basis for the processing.
2.3. Cooperation with data processors and third parties
If, in the course of our processing, we disclose data to other persons and companies (processors or third parties), transmit it to them or grant them access to the data, this will only be done on the basis of a legal permission (e.g. a transmission to payment service providers required for the performance of a contract pursuant to Art. 6 (1) lit. b GDPR), consent, a legal obligation or on the basis of our legitimate interests (e.g. when using agents, web hosts). If we commission third parties with the processing of data on the basis of a so-called "data processing agreement", this is done on the basis of Art. 28 GDPR.
2.4. Transfers to third countries
If we process data in a third country (i.e. outside the European Union or the European Economic Area) or if this is done in the context of using third-party services, this is only done if it is done to fulfil our (pre-)contractual obligations, on the basis of consent, due to a legal obligation or on the basis of our legitimate interests. We process data subject to legal or contractual permissions only if the requirements of Art. 44 et seq. GDPR are met: Data is therefore only processed if, for example, special guarantees exist, such as official recognition of a level of data protection corresponding to the EU (e.g. adequacy decision) or compliance with officially recognised contractual obligations (so-called standard contractual clauses).
2.5. Data deletion and storage period
As soon as the purpose of storage ceases to apply, personal data of a data subject will be deleted or its processing restricted ("blocked"). If European or national regulations, laws or other provisions to which we are subject require longer storage, the personal data will be stored in accordance with these legal provisions for longer than the original purpose intended.
2.6. Data deletion upon withdrawal
All data collected up to the point of withdrawal will be deleted immediately - unless a legal requirement requires retention.
3. Rights of the data subject
You have the right to gain access to the stored personal data.
You can obtain information from us about your stored data at any time (in accordance with Art. 15 GDPR).
You have the right to rectification.
You can request the correction of your data (in accordance with Art. 16 GDPR).
You have the right to erasure.
You can request the deletion (pursuant to Art. 17 of the GDPR) or a restriction of the processing of your data (pursuant to Art. 18 of the GDPR).
You have the right to data portability.
You can receive personal data that you have given us in a transferable format (in accordance with Article 20 of the GDPR).
You have the right to lodge a complaint.
You have the right to lodge a complaint with the competent supervisory authority (pursuant to Art. 77 DSGVO).
4. Data collection on our website
4.1. Server and log files
Our hosting provider collects the following data for statistical analysis: IP address of your computer at the time of access, date, time, retrieved content, the page from which you reached our homepage, operating system and browser.
4.2. Forms that require input (e.g. contact form)
You can fill out a contact form on our website. We store the data transmitted via the contact form from the input mask. We only process the data for the purpose of contacting you. The following data is collected: E-mail address, name, telephone number, and reason for your contact/message. The legal basis for processing the data that you transmit to us via the contact form is a (pre-)contractual obligation or your consent by transmitting your data for the purpose of contacting you. You can withdraw this consent at any time with effect for the future (informally by e-mail or post).
4.3. Mail
On our website you will find one or more e-mail addresses through which you can contact us. We store the personal data transmitted with the e-mail. We process the data only for the purpose of contacting you. The following data is collected: E-mail address, company, title, surname, first name, telephone number and reason for your contact. The legal basis for the processing of the data that you send us by e-mail is a (pre-)contractual obligation or your consent through the transmission of your data for the purpose of contacting you. You can revoke this consent at any time with effect for the future (informally by email or post).
5. Analysis and online marketing
Cookies are text files that are sent to your computer when you use websites in order to recognise it. Cookies are used to facilitate the use of websites and to make them more individual. You can set your web browser to inform you when cookies are being sent or to reject cookies. You can find information on this in the help function of your web browser (e.g. Firefox, Internet Explorer or Safari).
Our website does not use cookies or other tracking technologies.
6. Social Media: online presences and plug-ins
We maintain online presences in social networks and platforms in order to communicate with customers, interested parties and users active there and to inform them about our services. When accessing the respective networks and platforms, the terms and conditions and data processing guidelines of the respective operators apply.
7. Third-party services, tools and content
We do not use content or service offers from third parties to integrate their content and services (e.g. maps or fonts, hereinafter referred to as "content"). The prerequisite for this is that the third-party providers are aware of your IP address. We endeavour to only use content whose providers only use the IP address to deliver the content.
8. Contact with the data protection officer